How Is Dna A Template
If DNA is a book, then how is information technology read? Larn more virtually the Dna transcription procedure, where DNA is converted to RNA, a more portable set of instructions for the cell.
The genetic code is frequently referred to as a "blueprint" considering it contains the instructions a cell requires in guild to sustain itself. Nosotros now know that at that place is more to these instructions than simply the sequence of messages in the nucleotide code, nevertheless. For case, vast amounts of evidence demonstrate that this code is the footing for the production of various molecules, including RNA and poly peptide. Research has also shown that the instructions stored within DNA are "read" in ii steps: transcription and translation. In transcription, a portion of the double-stranded Dna template gives ascent to a single-stranded RNA molecule. In some cases, the RNA molecule itself is a "finished product" that serves some important role within the cell. Often, nonetheless, transcription of an RNA molecule is followed by a translation step, which ultimately results in the production of a protein molecule.
Visualizing Transcription
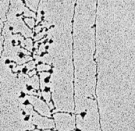
The procedure of transcription tin exist visualized by electron microscopy (Figure 1); in fact, it was showtime observed using this method in 1970. In these early electron micrographs, the DNA molecules appear every bit "trunks," with many RNA "branches" extending out from them. When DNAse and RNAse (enzymes that degrade DNA and RNA, respectively) were added to the molecules, the application of DNAse eliminated the trunk structures, while the use of RNAse wiped out the branches.
DNA is double-stranded, just but i strand serves every bit a template for transcription at any given time. This template strand is called the noncoding strand. The nontemplate strand is referred to equally the coding strand because its sequence will exist the same equally that of the new RNA molecule. In nearly organisms, the strand of Deoxyribonucleic acid that serves as the template for 1 cistron may be the nontemplate strand for other genes within the aforementioned chromosome.
The Transcription Process
The process of transcription begins when an enzyme called RNA polymerase (RNA politician) attaches to the template DNA strand and begins to catalyze production of complementary RNA. Polymerases are large enzymes composed of approximately a dozen subunits, and when agile on Dna, they are likewise typically complexed with other factors. In many cases, these factors signal which gene is to exist transcribed.
3 dissimilar types of RNA polymerase exist in eukaryotic cells, whereas bacteria take but i. In eukaryotes, RNA political leader I transcribes the genes that encode most of the ribosomal RNAs (rRNAs), and RNA politician Three transcribes the genes for one small rRNA, plus the transfer RNAs that play a key role in the translation process, as well as other pocket-size regulatory RNA molecules. Thus, information technology is RNA pol II that transcribes the messenger RNAs, which serve equally the templates for production of protein molecules.
Transcription Initiation

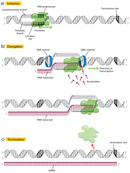
The kickoff step in transcription is initiation, when the RNA politico binds to the Dna upstream (5′) of the gene at a specialized sequence called a promoter (Figure 2a). In bacteria, promoters are usually composed of 3 sequence elements, whereas in eukaryotes, in that location are as many equally seven elements.
In prokaryotes, most genes have a sequence called the Pribnow box, with the consensus sequence TATAAT positioned about ten base pairs away from the site that serves as the location of transcription initiation. Not all Pribnow boxes have this exact nucleotide sequence; these nucleotides are only the most common ones found at each site. Although substitutions do occur, each box nonetheless resembles this consensus adequately closely. Many genes also take the consensus sequence TTGCCA at a position 35 bases upstream of the start site, and some have what is called an upstream element, which is an A-T rich region 40 to 60 nucleotides upstream that enhances the charge per unit of transcription (Figure 3). In any case, upon binding, the RNA political leader "core enzyme" binds to another subunit called the sigma subunit to form a holoezyme capable of unwinding the DNA double helix in order to facilitate access to the gene. The sigma subunit conveys promoter specificity to RNA polymerase; that is, information technology is responsible for telling RNA polymerase where to bind. At that place are a number of different sigma subunits that bind to different promoters and therefore aid in turning genes on and off as weather alter.
Eukaryotic promoters are more than complex than their prokaryotic counterparts, in part considering eukaryotes have the same three classes of RNA polymerase that transcribe different sets of genes. Many eukaryotic genes also possess enhancer sequences, which can exist found at considerable distances from the genes they affect. Enhancer sequences control gene activation past binding with activator proteins and altering the 3-D structure of the Dna to help "attract" RNA politico II, thus regulating transcription. Because eukaryotic DNA is tightly packaged equally chromatin, transcription too requires a number of specialized proteins that help make the template strand accessible.
In eukaryotes, the "cadre" promoter for a gene transcribed by politician 2 is most oft found immediately upstream (v′) of the get-go site of the factor. Most pol II genes have a TATA box (consensus sequence TATTAA) 25 to 35 bases upstream of the initiation site, which affects the transcription charge per unit and determines location of the outset site. Eukaryotic RNA polymerases use a number of essential cofactors (collectively called general transcription factors), and one of these, TFIID, recognizes the TATA box and ensures that the correct showtime site is used. Another cofactor, TFIIB, recognizes a different common consensus sequence, G/C One thousand/C One thousand/C K C C C, approximately 38 to 32 bases upstream (Figure 4).
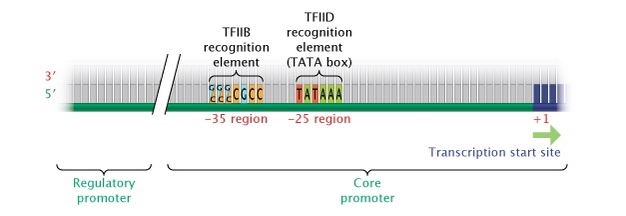
Effigy iv: Eukaryotic core promoter region.
In eukaryotes, genes transcribed into RNA transcripts by the enzyme RNA polymerase Ii are controlled past a cadre promoter. A core promoter consists of a transcription beginning site, a TATA box (at the -25 region), and a TFIIB recognition chemical element (at the -35 region).
© 2014 Nature Education Adjusted from Pierce, Benjamin. Genetics: A Conceptual Approach, 2nd ed. All rights reserved. ![]()
The terms "strong" and "weak" are often used to draw promoters and enhancers, according to their effects on transcription rates and thereby on gene expression. Alteration of promoter strength tin can have deleterious furnishings upon a jail cell, often resulting in illness. For example, some tumor-promoting viruses transform healthy cells past inserting strong promoters in the vicinity of growth-stimulating genes, while translocations in some cancer cells identify genes that should be "turned off" in the proximity of strong promoters or enhancers.
Enhancer sequences practice what their name suggests: They human action to enhance the rate at which genes are transcribed, and their effects can be quite powerful. Enhancers tin be thousands of nucleotides abroad from the promoters with which they interact, simply they are brought into proximity past the looping of Deoxyribonucleic acid. This looping is the result of interactions between the proteins bound to the enhancer and those bound to the promoter. The proteins that facilitate this looping are called activators, while those that inhibit information technology are chosen repressors.
Transcription of eukaryotic genes by polymerases I and III is initiated in a similar manner, simply the promoter sequences and transcriptional activator proteins vary.
Strand Elongation
Once transcription is initiated, the Dna double helix unwinds and RNA polymerase reads the template strand, calculation nucleotides to the 3′ stop of the growing concatenation (Figure 2b). At a temperature of 37 degrees Celsius, new nucleotides are added at an estimated rate of about 42-54 nucleotides per 2nd in leaner (Dennis & Bremer, 1974), while eukaryotes go along at a much slower pace of approximately 22-25 nucleotides per second (Izban & Luse, 1992).
Transcription Termination
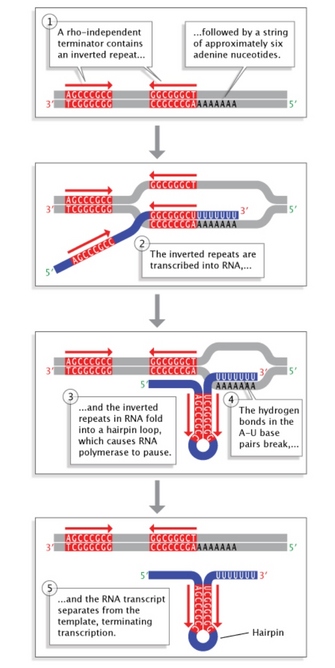
Effigy 5: Rho-contained termination in bacteria.
Inverted echo sequences at the terminate of a gene allow folding of the newly transcribed RNA sequence into a hairpin loop. This terminates transcription and stimulates release of the mRNA strand from the transcription mechanism.
© 2014 Nature Education Adapted from Pierce, Benjamin. Genetics: A Conceptual Approach, 2nd ed. All rights reserved. ![]()
Terminator sequences are plant close to the ends of noncoding sequences (Figure 2c). Bacteria possess two types of these sequences. In rho-independent terminators, inverted repeat sequences are transcribed; they can then fold back on themselves in hairpin loops, causing RNA pol to pause and resulting in release of the transcript (Effigy v). On the other hand, rho-dependent terminators brand use of a factor called rho, which actively unwinds the DNA-RNA hybrid formed during transcription, thereby releasing the newly synthesized RNA.
In eukaryotes, termination of transcription occurs by different processes, depending upon the exact polymerase utilized. For politician I genes, transcription is stopped using a termination factor, through a mechanism like to rho-dependent termination in bacteria. Transcription of political leader III genes ends subsequently transcribing a termination sequence that includes a polyuracil stretch, by a machinery resembling rho-independent prokaryotic termination. Termination of pol II transcripts, however, is more complex.
Transcription of politician II genes can go on for hundreds or fifty-fifty thousands of nucleotides beyond the end of a noncoding sequence. The RNA strand is so broken past a complex that appears to associate with the polymerase. Cleavage seems to be coupled with termination of transcription and occurs at a consensus sequence. Mature politico II mRNAs are polyadenylated at the iii′-end, resulting in a poly(A) tail; this process follows cleavage and is also coordinated with termination.
Both polyadenylation and termination brand use of the aforementioned consensus sequence, and the interdependence of the processes was demonstrated in the belatedly 1980s by piece of work from several groups. One group of scientists working with mouse globin genes showed that introducing mutations into the consensus sequence AATAAA, known to exist necessary for poly(A) addition, inhibited both polyadenylation and transcription termination. They measured the extent of termination past hybridizing transcripts with the different poly(A) consensus sequence mutants with wild-type transcripts, and they were able to see a decrease in the signal of hybridization, suggesting that proper termination was inhibited. They therefore ended that polyadenylation was necessary for termination (Logan et. al., 1987). Another group obtained similar results using a monkey viral organisation, SV40 (simian virus 40). They introduced mutations into a poly(A) site, which caused mRNAs to accumulate to levels far to a higher place wild type (Connelly & Manley, 1988).
The exact relationship between cleavage and termination remains to exist determined. One model supposes that cleavage itself triggers termination; another proposes that polymerase activity is affected when passing through the consensus sequence at the cleavage site, perhaps through changes in associated transcriptional activation factors. Thus, research in the area of prokaryotic and eukaryotic transcription is still focused on unraveling the molecular details of this circuitous process, data that will allow us to improve understand how genes are transcribed and silenced.
References and Recommended Reading
Connelly, Southward., & Manley, J. 50. A functional mRNA polyadenylation bespeak is required for transcription termination by RNA polymerase Ii. Genes and Evolution four, 440–452 (1988)
Dennis, P. P., & Bremer, H. Differential rate of ribosomal poly peptide synthesis in Escherichia coli B/r. Journal of Molecular Biology 84, 407–422 (1974)
Dragon. F., et al. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417, 967–970 (2002) doi:10.1038/nature00769 (link to article)
Izban, M. G., & Luse, D. Southward. Factor-stimulated RNA polymerase Ii transcribes at physiological elongation rates on naked DNA but very poorly on chromatin templates. Periodical of Biological Chemistry 267, 13647–13655 (1992)
Kritikou, E. Transcription elongation and termination: It ain't over until the polymerase falls off. Nature Milestones in Gene Expression eight (2005)
Lee, J. Y., Park, J. Y., & Tian, B. Identification of mRNA polyadenylation sites in genomes using cDNA sequences, expressed sequence tags, and trace. Methods in Molecular Biology 419, 23–37 (2008)
Logan, J., et al. A poly(A) add-on site and a downstream termination region are required for efficient cessation of transcription past RNA polymerase Ii in the mouse beta maj-globin gene. Proceedings of the National University of Sciences 23, 8306–8310 (1987)
Nabavi, S., & Nazar, R. N. Nonpolyadenylated RNA polymerase II termination is induced by transcript cleavage. Periodical of Biological Chemistry 283, 13601–13610 (2008)
How Is Dna A Template,
Source: https://www.nature.com/scitable/topicpage/dna-transcription-426/
Posted by: saucierdring1986.blogspot.com


0 Response to "How Is Dna A Template"
Post a Comment